Overview
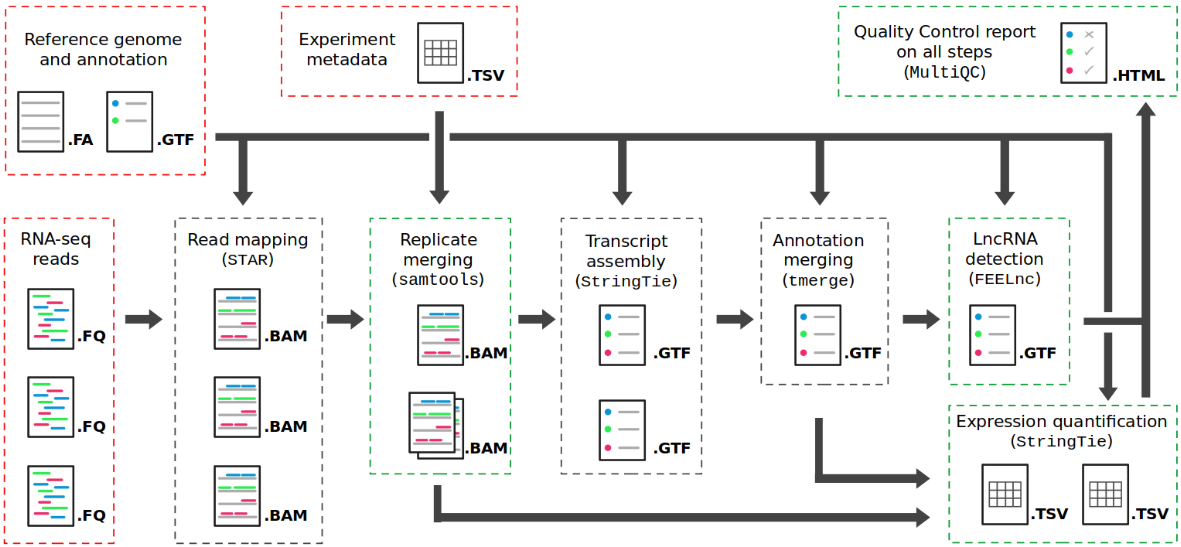
TAGADA is an RNA-seq data analysis pipeline for Transcript And Gene Assembly, Deconvolution, and Analysis. From a genomic sequence, a reference gene annotation and RNA-seq reads, TAGADA can improve the reference annotation by finding new genes and transcripts. Gene expresion is quantified for genes and transcripts of both the reference and the extended annotations. Long non-coding transcripts are detected. TAGADA is easy to use, portable, customizable, and provides a detailed Quality Control report. TAGADA is implemented in NextFlow DSL2 and is available as a Docker or Singularity container. It has been developped to profile transcriptomic maps of the pig and chicken genomes during development in the context of the GENE-SWitCH project (7 tissues, 3 developmental stages, 4 animals).
Resources
Source code
TAGADA's code is open-source and freely available on the FAANG Github repository.
Article and additional data
The first submitted version of the manuscript along with complementary data are available at the INRAE Omics dataverse repository.
Funding
Reference
If you use this resource, please cite the following article:
TAGADA: a scalable pipeline to improve genome annotations with RNA-seq data.
Kurylo C, Guyomar C, Foissac S, Djebali S (2023)
Submitted
DOI: 10.57745/3UGLXW



